
GROMACS
GROMACS is a toolkit to perform molecular dynamics simulations of biomolecules using Newton’s equations of motion. According to the Stanford 2018 HPC update, it is the world’s most widely open source HPC application accounting for 5% of worldwide supercomputing resources, and according to the NVIDIA-commissioned Intersect360 survey it ranks #1 across all fields (academic and commercial) for GPU computing applications, and has over 40,000 citations.
MCP: SESSI

OpenSpace
OpenSpace is an open-source astrovisualization tool that aims at bridging the gap between the domain scientists and the general public to portray our efforts in exploring the cosmos and showing these efforts in their proper context. OpenSpace runs on standard computer hardware and dome planetariums and also can be used by scientists in their data analysis which makes it possible to increase public outreach opportunities. In addition, OpenSpace enables simultaneous connections across the globe creating opportunity for shared experiences among audiences worldwide. OpenSpace is open source, non-commercial, and freely available software that is enabled through the generous support from the Swedish e-Science Research Centre, NASA, and the Knut and Alice Wallenberg Foundation. The project stems from the same academic collaboration between Sweden’s Linköping University (LiU) and the American Museum of Natural History (AMNH).
NEK5000
KTH-Framework for 3rd party Nek5000 tools
We developed a framework for 3rd party tools that could be used with SEM code Nek5000. The goal was to provide a platform for easy development and sharing of code built on top of Nek5000. There are multiple packages performing various tasks available in the toolbox right now. Some of them are relatively simple (e.g. mpi-rank independent white noise generator), some other quite advanced (e.g. eigenvalue calculation using Arnoldi algorithm). One of the main features of the framework is code re-usability, which allows to build new tools based on the existing ones. An example is turbulent statistics tool, which uses more general 3D to 2D mesh mapping package. An important part of the toolbox is a set of examples and simple instructions describing usage of given code package. Although most of the tools are developed at KTH, it is used by number of Nek5000 users at different countries.
https://github.com/KTH-Nek5000/KTH_Framework
A wavemaker: an overlap between direct and adjoint strongest eigenmodes for jet in cross-flow simulation.
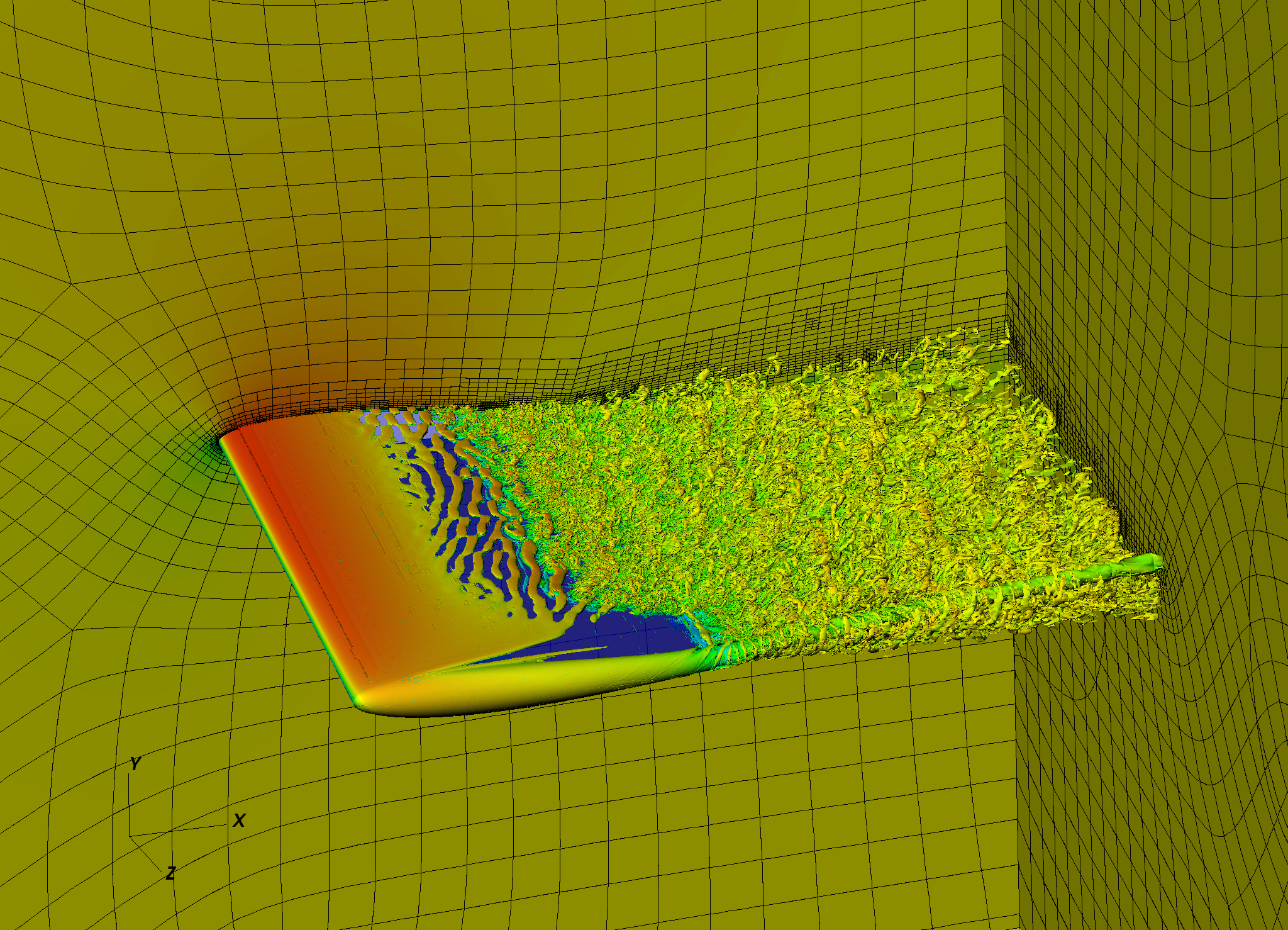
Adaptive Mesh Refinement version of SEM solver Nek5000
A quality and performance of a high fidelity CFD modelling depends strongly on the mesh used for a simulation. The goal is to generate a mesh that with minimum amount of degrees of freedom would provide smallest possible computational error. This can be achieved in a number of ways, although the prior knowledge of the flow dynamics is required. We implemented Adaptive Meh Refinement algorithm in SEM solver Nek5000 allowing to build optimal mesh without in-depth knowledge of the flow. It uses estimate of the computational error for a given mesh structure and performs dynamical mesh modification during the simulation to minimise the error. We utilise h-type refinement which allows to locally increase number of degrees in the regions with high computational error. Repeating this process leads to the mesh that is close to optimal and provides high resolution at the position where it is really needed. Such algorithms are essential for industrially-relevant simulations of turbulent flows, that otherwise would not be affordable.
VeloxChem
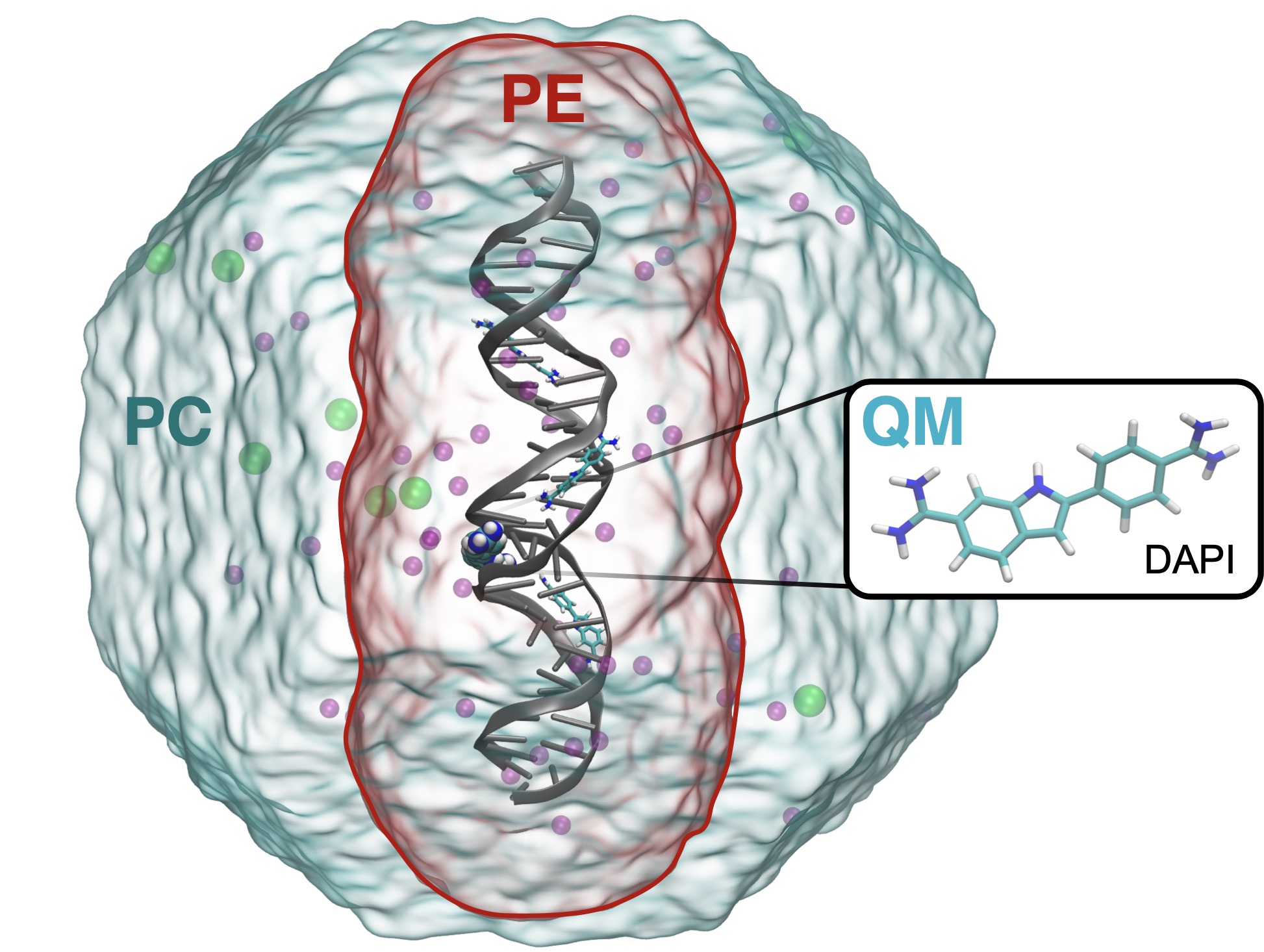
VeloxChem is an open-source program for the calculation of electronic real and complex linear response functions at the levels of Hartree–Fock and Kohn–Sham density functional theory. With an object-oriented program structure written in a Python/C++ layered fashion, VeloxChem enables time-efficient prototyping of novel scientific approaches without sacrificing computational efficiency, so that molecular systems involving up to and beyond 500 second-row atoms (or some 10,000 contracted and in part diffuse Gaussian basis functions) can be routinely addressed. In addition, VeloxChem is equipped with a polarizable embedding scheme for the treatment of the classical electrostatic interactions with an environment that in turn is modeled by atomic site charges and polarizabilities.
The underlying hybrid MPI/OpenMP parallelization scheme makes VeloxChem suitable for execution in high-performance computing (HPC) cluster environments, showing even slightly beyond linear scaling for the Fock matrix construction with use of up to 20,000 CPU cores. An efficient—with respect to convergence rate and overall computational cost—multi-frequency/gradient complex linear response equation solver enables calculations not only of conventional spectra, such as visible/UV/X-ray electronic absorption and circular dichroism spectra, but also time-resolved linear response signals as due to ultra-short weak laser pulses. VeloxChem is distributed under the GNU Lesser General Public Licence version 2.1 (LGPLv2.1) licence and made available for download from the homepage https://veloxchem.org.
Relevant publication:
Rinkevicius, Z.; Li, X.; Vahtras, O.; Ahmadzadeh, K.; Brand, M.; Ringholm, M.; List, N. H.; Scheurer, M.; Scott, M.; Dreuw, A.; Norman, P. VeloxChem: A Python‐driven Density‐functional Theory Program for Spectroscopy Simulations in High‐performance Computing Environments. WIREs Comput. Mol. Sci. 2020, 10 (5), 1–14. https://doi.org/10.1002/wcms.1457.
MCP: DCMD
Gator
The Gator program is available for computational spectroscopy and calculations of molecular properties using real and complex propagators at the correlated level of wave function theory. Currently, the focus lies on methods based on the algebraic diagrammatic construction (ADC) scheme up to the third order of perturbation theory. An auxiliary Fock matrix-driven implementation of the second-order ADC method for excitation energies has been realized with an underlying hybrid MPI/OpenMP parallelization scheme suitable for execution in high-performance computing cluster environments. VeloxChem is distributed under the GNU Public Licence version 3 (GPLv3) licence and made available for download from the homepage https://github.com/gator-program/gator.
Relevant publication:
(1) Rehn, D. R.; Rinkevicius, Z.; Herbst, M. F.; Li, X.; Scheurer, M.; Brand, M.; Dempwolff, A. L.; Brumboiu, I. E.; Fransson, T.; Dreuw, A.; Norman, P. Gator: A Python‐driven Program for Spectroscopy Simulations Using Correlated Wave Functions. WIREs Comput. Mol. Sci. 2021. https://doi.org/10.1002/wcms.1528.
MCP: DCMD
NEP-PACK: Nonlinear eigenvalue problem algorithms package
NEP-PACK is a novel open-source Julia language library for the solution of nonlinear eigenvalue problems (NEPs). The package provides a framework to represent NEPs, as well as efficient implementations of many state-of-the-art linear algebra algorithms for nonlinear eigenvalue problems, including many important algorithm classes such as Krylov methods, preconditioning methods, contour integral methods, deflation methods and Jacobi-Davidson methods. The package is designed for high-performance computing users and application researchers as well as algorithm developers. The data structures are designed with applications and algorithms in mind such that a large range of efficient solvers can be used for a wide range of problems, including problems not only defined in Julia, but also MATLAB, Python, Fortran and C. Tutorials and examples illustrate how to use the package to solve problems arising in many settings and fields, e.g., quantum physics, nanophotonics, fluid mechanics, etc.
https://github.com/nep-pack/NonlinearEigenproblems.jl
MCP: Data Science
Simson
Simson version 4.0.0 is a software package that implements an efficient spectral integration technique to solve the Navier–Stokes equations for incompressible channel and boundary layer flows. The report describes how to configure, compile and install the software. Additionally, an introduction to the theory and the numerical details of the implementation is given. The solver is implemented in Fortran 77/90. The original algorithm reported in Lundbladh et al. (1992a) solved the incompressible Navier–Stokes equations in a channel flow geometry. That algorithm has been reimplemented in a boundary layer version of the code reported in Lundbladh et al. (1999). That allowed simulations of the flow over a flat plate. To do this an artificial free-stream boundary condition was introduced, and for spatial simulations a fringe region technique similar to that of Bertolotti et al. (1992) was implemented. In Simson the channel and boundary layer solvers have been combined together with many different features developed over the years. The code can be run either as a solver for direct numerical simulation (DNS) in which all length and time scales are resolved, or in large-eddy simulation (LES) mode where a number of different subgrid-scale models are available. The evolution of multiple passive scalars can also be computed. The code can be run with distributed or with shared memory parallelization through the Message Passing Interface (MPI) or OpenMP, respectively. The wall-parallel directions are discretized using Fourier series and the wall-normal direction using Chebyshev series. Time integration is performed using a third order Runge–Kutta method for the advective and forcing terms and a Crank–Nicolson method for the viscous terms. The basic numerical method is similar to the Fourier–Chebyshev method used by Kim et al. (1987). Further details about spectral discretizations and additional references are given in e.g. Canuto et al. (1988).
GitHub repository: https://github.com/KTH-Nek5000/SIMSON
Relevant publication:
Spectral Simulations of Wall-bounded Flows on Massively-parallel Computers (Qiang Li, Philipp Schlatter, Dan Henningson, 2008): li_schlatter_henningson_2008.pdf
SIMSON- A Pseudo-Spectral Solver for Incompressible Boundary Layer Flows (Mattias Chevalier, Philipp Schlatter, Anders Lundbladh, Dan Henningson): chevalier_schlatter_lundbladh_henningson_2007.pdf
MCP: SESSI

UQit
UQit is an open-source Python package for uncertainty quantification (UQ) in computational physics, in general, and computational fluid dynamics (CFD), in particular. Currently, different UQ techniques are available in the package for uncertainty propagation, global sensitivity analysis, and surrogate construction. Due to the non-intrusive nature of the implemented UQ techniques, UQit can be linked to any simulator (e.g. CFD solver) conditioned on having appropriate interfaces. The flexible design of UQit together with the fact that Python is rich with data science libraries, provide a good potential for further development of UQit in response to different needs coming up in particular applications.
GitHub repository: https://github.com/KTH-Nek5000/UQit
PyPI repository: https://pypi.org/project/UQit/
Relevant publication:
Rezaeiravesh, S., Vinuesa, R., & Schlatter, P. (2020). An uncertainty-quantification framework for assessing accuracy, sensitivity, and robustness in computational fluid dynamics. arXiv:2007.07071. https://doi.org/arXiv:2007.07071
MCP: SESSI
UppASD
The UppASD package is a simulation tool for atomistic spin dynamics at finite temperatures. The program evolves in time the equations of motion for atomic magnetic moments in a solid. The equations take the form of the Landau-Lifshitz-Gilbert (LLG) equation. For most of the applications done so far, the magnetic exchange parameters of a classical Heisenberg Hamiltonian have been used in ASD simulations. The parameters are extracted from ab-initio DFT codes.
Within the SeRC activities, the focus is to develop a combined spin-dynamics and molecular dynamics tool within UppASD.
https://physics.uu.se/forskning/materialteori/pagaende-forskning/uppasd/
MCP: DCMP

Inviwo
Inviwo is an open-source visualization software developed by a consortium led by Linköping University partnered with Ulm University and KTH. Inviwo is a software framework for the rapid prototyping of visualizations. It is written in C++, exploits modern graphics hardware, and is available under BSD license, which permits free use in any setup – also commercially.
Website: https://inviwo.org
Github: https://github.com/inviwo/inviwo

climt
climt is a toolkit for building Earth system models in Python. climt stands for Climate Modelling and Diagnostics Toolkit — it is meant both for creating models and for generating diagnostics (radiative fluxes for an atmospheric column, for example).
https://github.com/CliMT/climt
Community: Climate Modeling
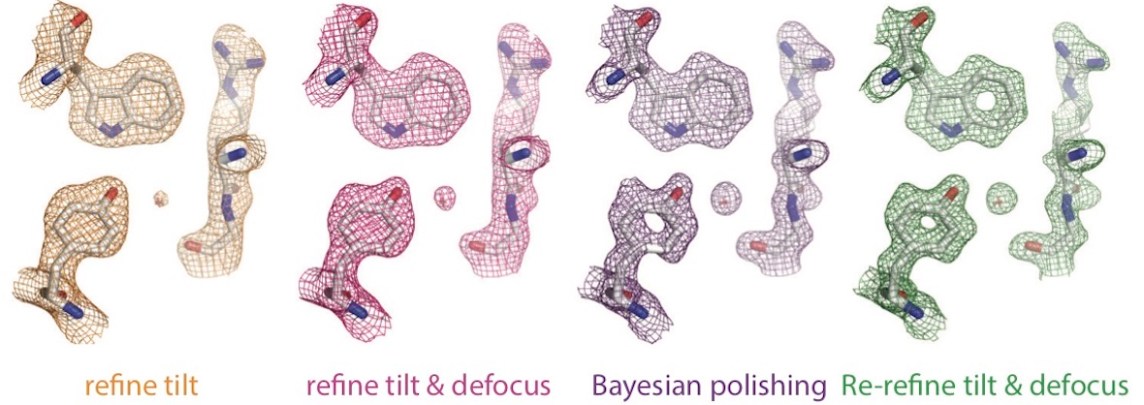
RELION
RELION is a code used for the refinement of macromolecular structures from single-particle cryo-electron microscopy data, and with the recent revolution in terms of high resolution cryo-EM, it has become a central tool in structural biology with several thousand citations. The code is developed as a collaboration between MRC-LMB and Science for Life Laboratory.
Example reference: Kimanius, D., Forsberg, BO., Scheres, SHW., Lindahl, E. (2016) Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722.
https://www3.mrc-lmb.cam.ac.uk/relion/index.php
MCP: SESSI
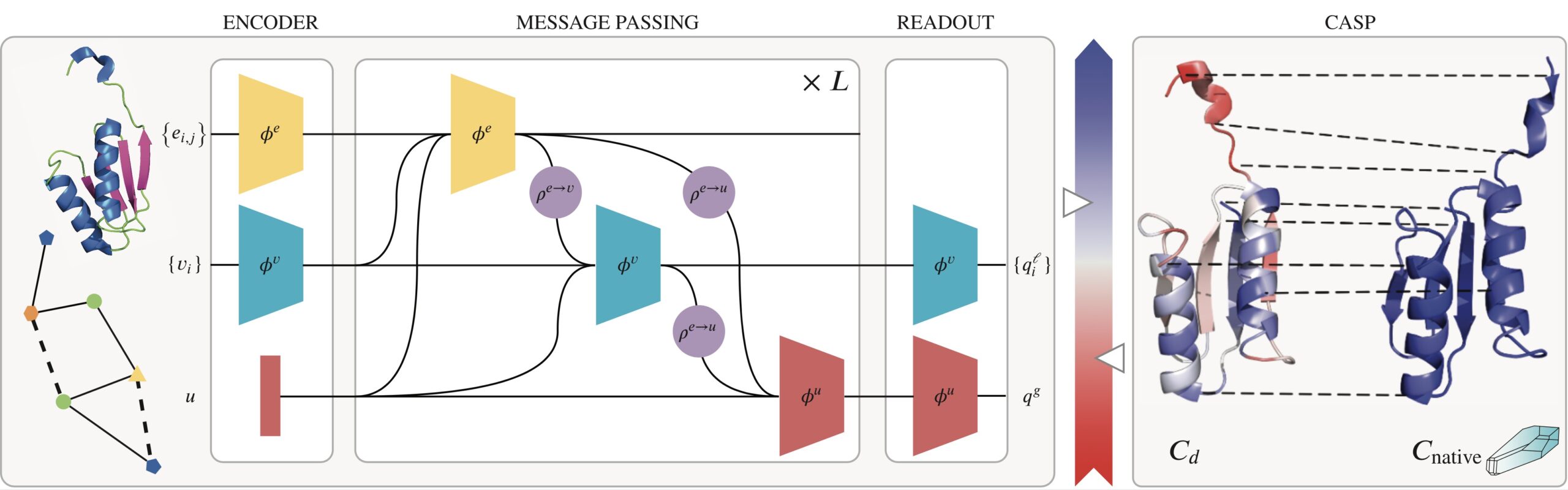
GraphQA
The opensource project concerns protein model quality assessment as part of the Data Science MCP. It consists of feature extraction, training and evaluation. The method is based on a deep learning technique called Graph Convolutional Networks (GCN) that can operate on graphs as input. As such, proteins are represented as graphs with hand-designed and/or learned features for both nodes (amino-acides) and edges (chemical and/or 3D location relations). The model is trained using local and global loss functuons based on native protein models. The method has also entered the CASP14 challenge on protein model accuracy.
Refference paper: Baldassarre et al. “GraphQA: Protein Model Quality Assessment using Graph Convolutional Networks”, Bioinformatics 2020 (https://doi.org/10.1093/bioinformatics/btaa714)
GitHub Link: https://github.com/baldassarreFe/graphqa
Server Link: http://isengard.csc.kth.se:8585/
MCP: Data Science

The MISU MIT Cloud and Aerosol (MIMICA) model
MIMICA is a large-eddy simulation code designed to study cloudy planetary boundary layers (Savre et al., 2014). The model solves the non-hydrostatic anelastic equations using high-order low-dissipative numerical schemes for the advection of scalars and momentum. MIMICA contains a two-moment bulk microphysics scheme representing five types of hydrometeors including ice crystals and snow. In addition, an aerosol module can be dynamically
coupled to the solver providing the possibility for a detailed representation of cloud-aerosol feedbacks. The model has been used for numerous studies of sub-tropical and Arctic clouds. It is currently also being developed and used for deep convective cloud studies.
Reference paper: Savre J., A. M. L. Ekman, and G. Svensson (2014), Technical note: Introduction to MIMICA, a large-eddy simulation solver for cloudy planetary boundary layers, J. Adv. Model. Earth Syst., 6, doi:10.1002/2013MS000292.
https://bitbucket.org/matthiasbrakebusch/mimicav5/src/master/ (requires permission which can be obtained by contacting matthias.brakebush@aces.su.se)
Community: Climate Modeling
Web-server for TOPCONS2
This is the web-server implementation of the TOPCONS2 workflow. The web-server is developed with Django 2.2.7+ and Python 3.6+. This software is open source and licensed under the MIT license.
TOPCONS2 is an updated version of the widely used TOPCONS for predicting membrane protein topologies using consensus prediction. It is faster yet more accurate than the old TOPCONS according to our solid benchmarking. Moreover, it predicts not only the trans-membrane helices, but also the location of signal peptide
This implementation employs two queuing schemes for small jobs and large jobs respectively. For single-sequence jobs submitted via web-page, they will be run directly (and usually immediately after submission) at the front-end server. For multiple-sequence jobs or jobs submitted via the API (a Python script for the command-line use of the API is included in the package), they will be forwarded to the remote servers via the WSDL (Web Service Definition Language) service. Consequently, the web-server can handle jobs of all proteins from a proteome. This implementation is suitable as as a base platform for bioinformatic prediction tools that need to be run for one or many sequences but the computational time for each sequence is short.
https://github.com/ElofssonLab/web_topcons2
MCP: Data Science
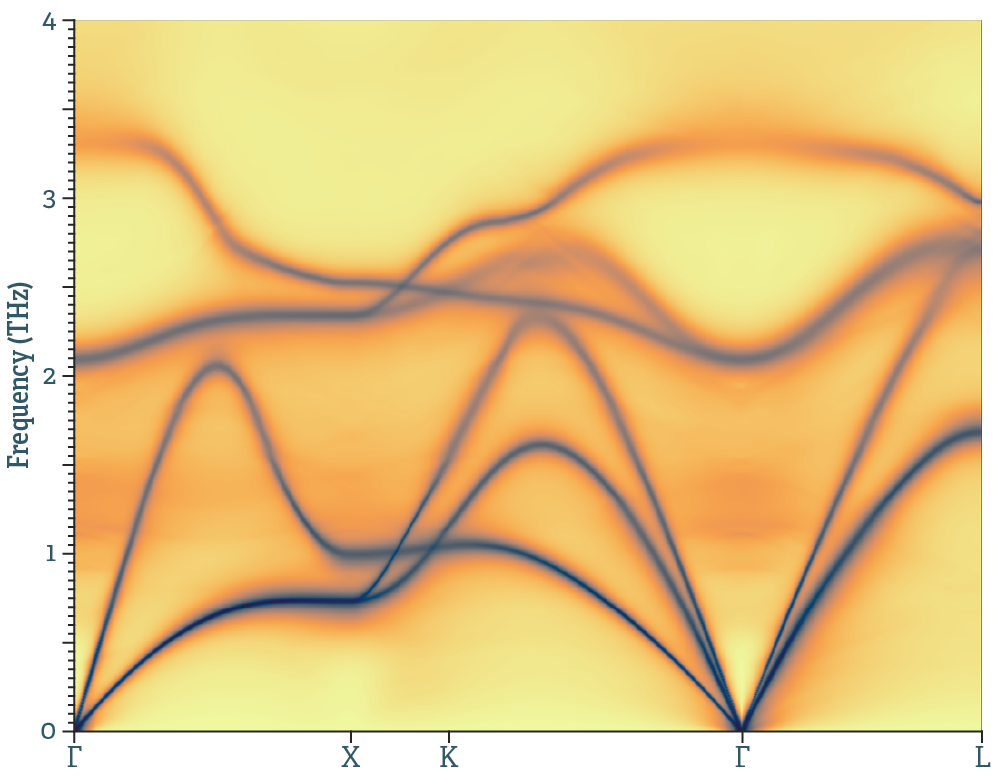
Temperature dependent effective potential
A package that deals with finite temperature lattice dynamics in solids. Capabilities include, but are not limited to, vibrations in strongly anharamonic systems, inelastic neutron scattering, thermal transport and thermodynamics of materials.
MCP: DCMD
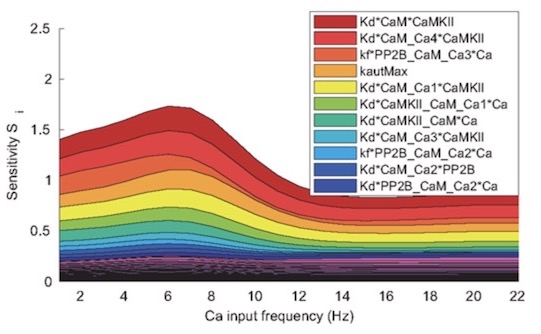
Uncertainty Quantification (UQ) and Sensitivity Analysis (SA)
The code distributed in this repository implements the methodology presented in the paper “Uncertainty quantification, propagation and characterization by Bayesian analysis combined with global sensitivity analysis applied to dynamical intracellular pathway models” by Eriksson & Jauhiainen et al (2018). The code is distributed under the GNU General Public License v3.0.
https://github.com/alexjau/uqsa
MCP: Brain-IT
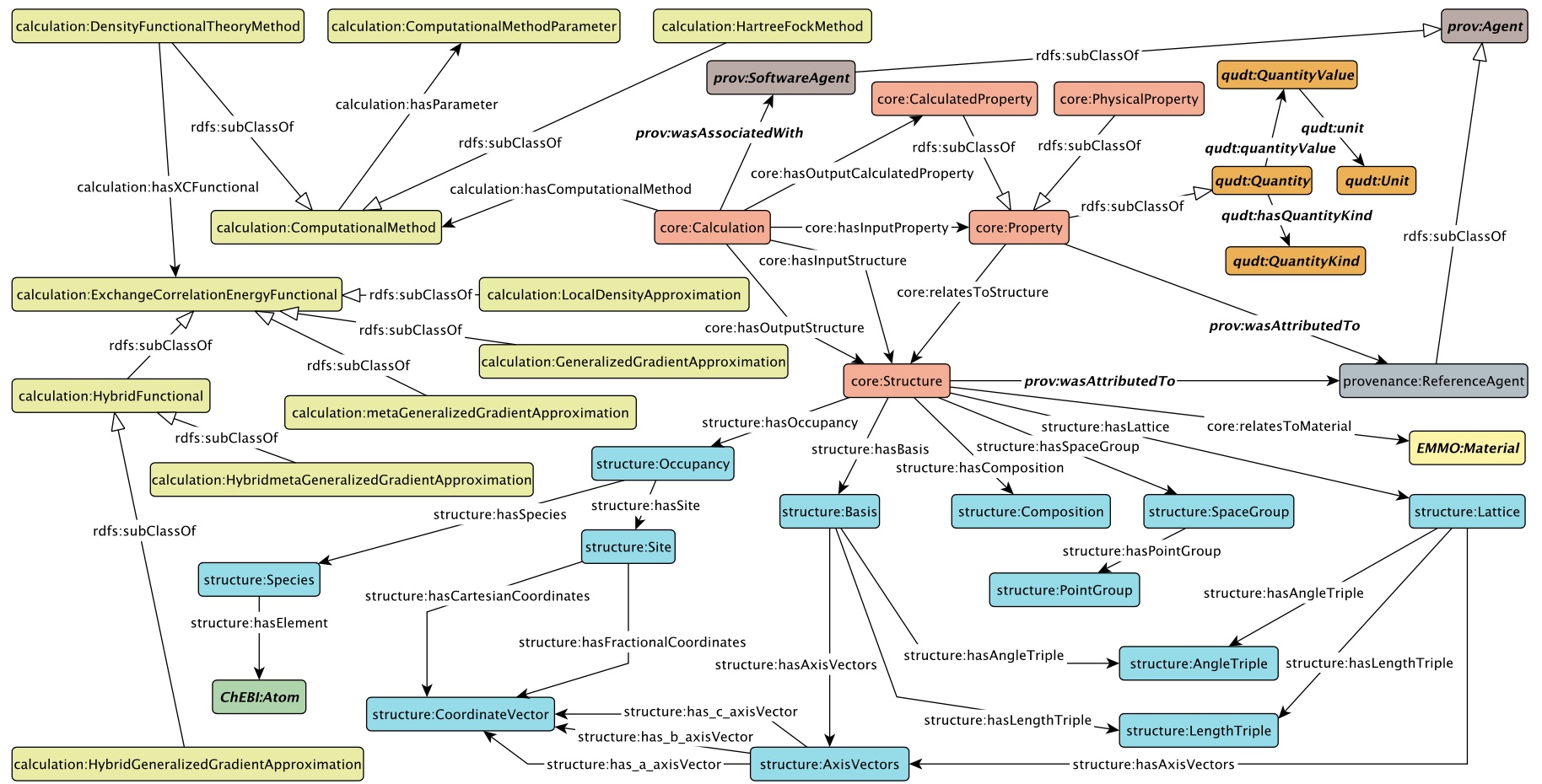
Materials Design Ontology (MDO)
MDO is an ontology for the materials design field, representing domain knowledge specifically related to solid-state physics and computational materials science. Ontologies are used in the process of making data Findable, Accessible, Interoperable and Reusable (FAIR). MDO is such a resource for materials databases and was guided by the work in the OPTIMADE (https://www.optimade.org/) consortium.
Reference paper: Huanyu Li, Rickard Armiento, Patrick Lambrix (2020), An Ontology for the Materials Design Domain (doi: https://doi.org/10.1007/978-3-030-62466-8_14)
Link: https://w3id.org/mdo
MCP: DCMD

percMPI: Percolation for node-distributed data
The percMPI repository contains the code for a novel memory-distributed parallel algorithm to finely sample the percolation function. THe algorithm is based on a parallel version of the union-find algorithm interleaved with a global synchronization step for each threshold sample. The efficiency of this algorithm stems from the fact that operations in-between threshold samples can be freely reordered, are mostly local and thus require no inter-process communication. The algorithm is significantly faster than previous algorithms, and is neither constrained by memory size nor number of compute nodes.
Relevant reference: Anke Friederici*, Wiebke Köpp*, Marco Atzori, Ricardo Vinuesa, Philipp Schlatter, Tino Weinkauf. Distributed Percolation Analysis for Turbulent Flows. 9th IEEE Symposium on Large Data Analysis and Visualization (LDAV), 2019. DOI: 10.1109/LDAV48142.2019.8944383, links to data sets and videos.
*Both authors contributed equally

In-situ analysis in Catalyst and Nek5000 with Mesa
We describe our implementation of an in-situ adaptor for Nek5000 and ParaView/Catalyst, as well the building process of all its components, and a test case. The test case is the simulation of an incompressible flow around a NACA4412 airfoil. The setup of the test case partially replicates that of a realistic high-fidelity simulation, but we employed a sufficiently coarse resolution to allow testing on a standard personal computer.
Relevant reference: Wiebke Köpp, Marco Atzori, Mohamad Rezaei, Niclas Jansson, Ricardo Vinuesa, Erwin Laure, Philipp Schlatter, and Tino Weinkauf. Effects of Blowing and Suction on the Turbulent Flow around an Airfoil. 73rd Annual Meeting of the APS Division of Fluid Dynamics. Gallery of Fluid Motion 2020. DOI: 10.1103/APS.DFD.2020.GFM.V0058

R Packages Developed Through eCPC
Microsimulation and Prostata
microsimulation is an R package for discrete event simulation in R and C++. It includes facilities for common random numbers and in-simulation reporting, particularly for health economic evaluations. The prostata package extends the microsimulation package to model for prostate cancer screening. The prostata package has been well validated for Sweden and for the European Randomised Study of Prostate Cancer.
Example reference: Karlsson et al (https://doi.org/10.1109/eScience.2016.7870915; https://doi.org/10.1371/journal.pone.0211918)
https://github.com/mclements/microsimulation
rstpm2
R implementation of generalized survival models (GSMs), smooth accelerated failure time (AFT) models and Markov multi-state models.
Example reference: Liu et al (https://doi.org/10.1177/0962280216664760)
magree
Implements the O’Connell-Dobson-Schouten Estimators of Agreement for Multiple Observers
Example reference: Egevad et al (https://doi.org/10.1007/s00428-020-02858-w)
biostat3
Utility functions, datasets and extended examples for survival analysis. This extends a range of other packages, some simple wrappers for time-to-event analyses, datasets, and extensive examples in HTML with R scripts. The package also supports the course Biostatistics III entitled “Survival analysis for epidemiologists in R”.




