Projects

Large-scale ice sheet modeling
We focus on improving the efficiency of ice sheet models coupled to the ocean. Constructing accurate models of the ice sheets on Greenland and Antarctica is crucial in order to reliably estimate sea level rise. Due to low efficiency of todays models it is currently not possible to run accurate enough simulations for long enough time spans.
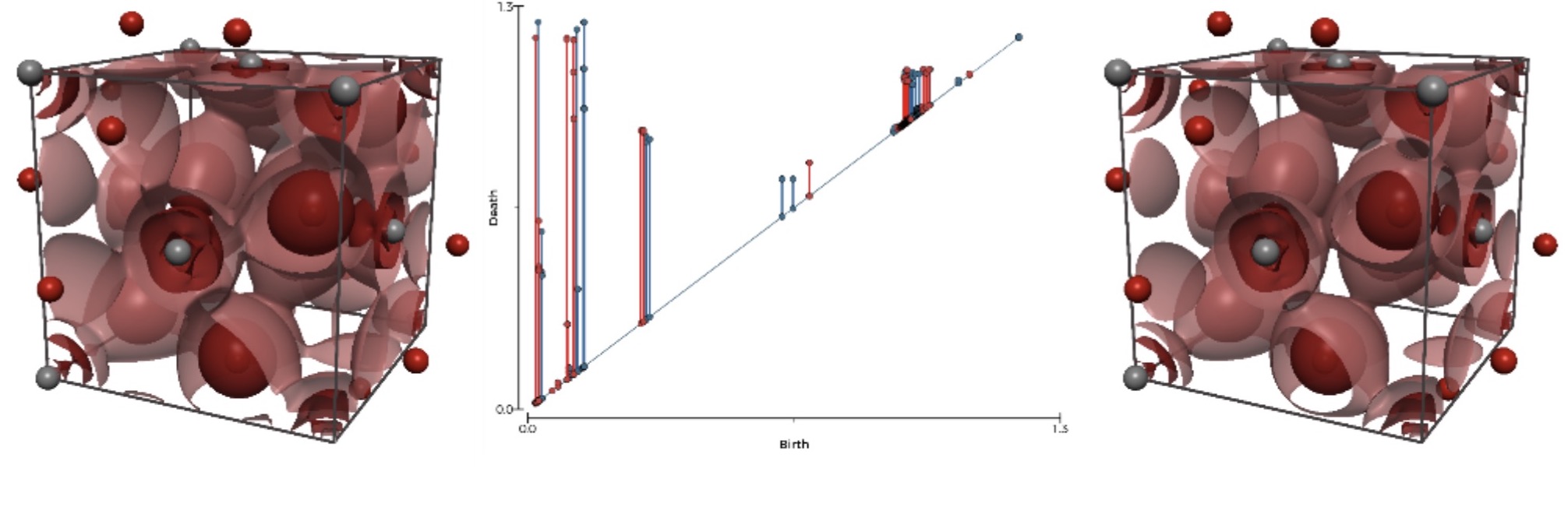
Data exploration and visualization
Method development is needed into data exploration and visualization of the generated materials data. This is a new and exciting field of multidisciplinary research where data science meets computational materials physics. Specific activities in this group include:

Automatic workflow, data collection, and development of open-data infrastructure
For DCMD activities in data exploration and visualization, we need to address generation, collection, storing, and organizing data via research on automatic workflow, data collection for materials data, and development of an open-data infrastructure. All activities lead to necessary insights and software to do complex simulations within materials physics and molecular chemistry.
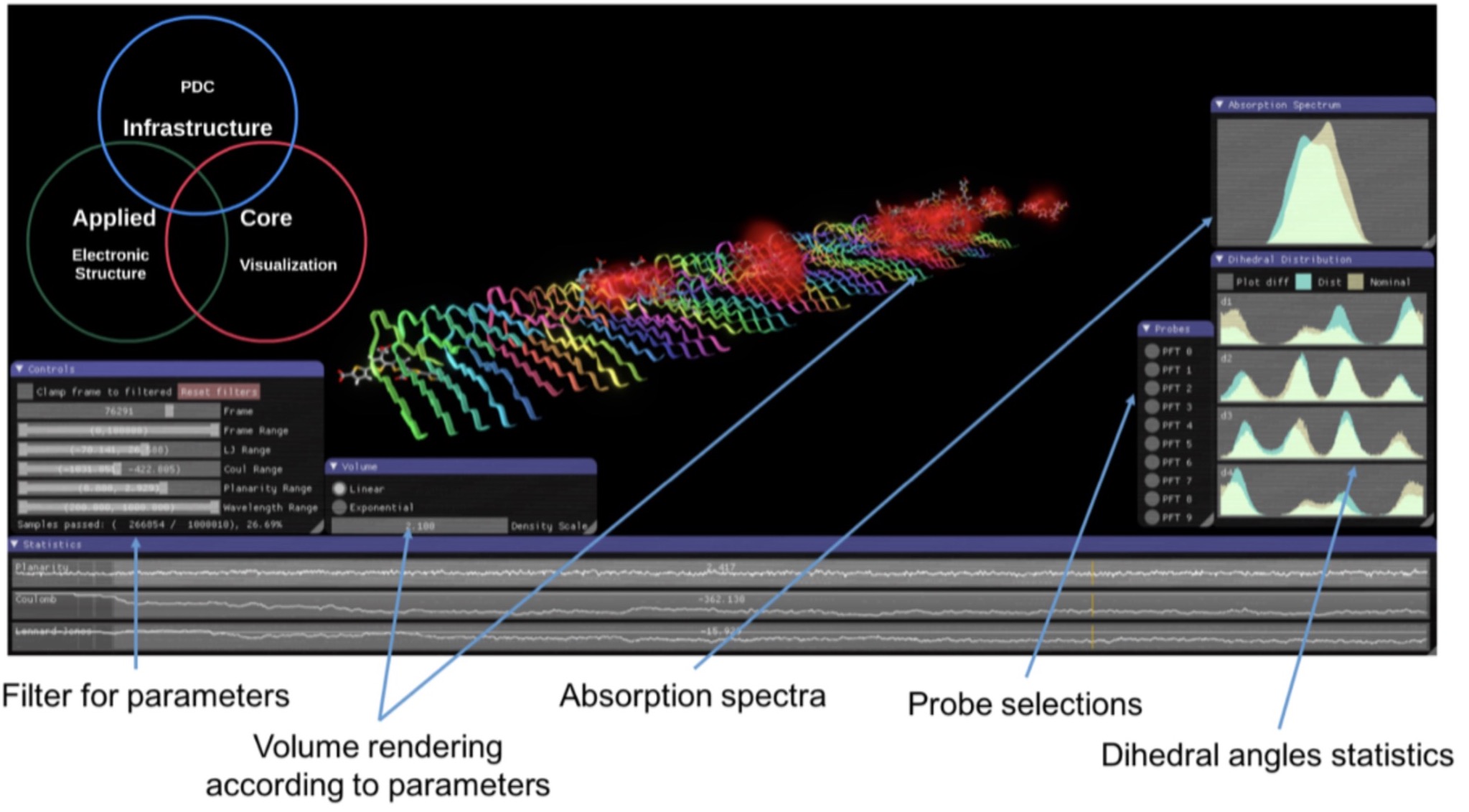
Software development for exploration and design of complex molecular systems
The dominating software for quantum molecular simulations is an American commercial product (Gaussian). In an undertaking together with PDC, we will develop a full-fledged DFT program with all the standard capabilities as well as non-standard functionalities developed in the Scandinavian Dalton program community and which provides state-of-the-art scaling on contemporary and future HPC hardware platforms based on Intel, ARM, and Power CPUs as well as NVIDIA GPUs.
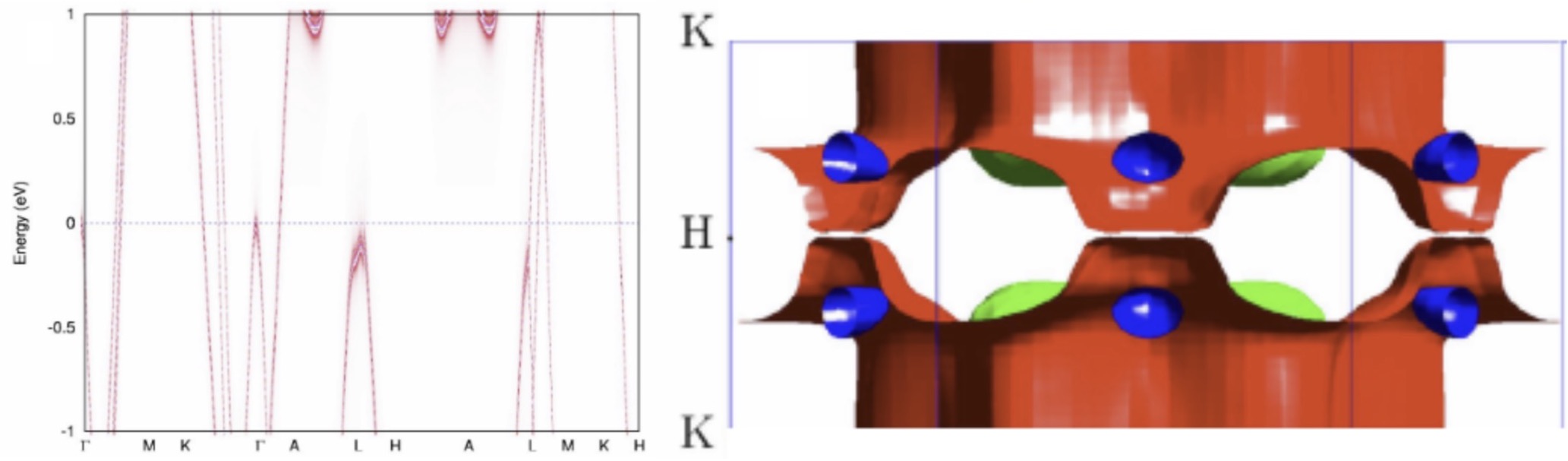
Development of novel modeling techniques
A need for high quality large volume materials data requires basic research into thedevelopment of novel modeling techniques. This work concerns method development with increased accuracy and efficiency, including dynamical mean-field theory (DMFT), spin- dynamics, time-dependent response theory (TDRSP), and molecular dynamics (MD).

AI-centered visual analytics of histology
This project aims to merge AI techniques with visual exploration.

Organic materials for the future
Atomistic MD simulations of polymer- and cellulose-based materials are performed to investigate the impact of ionic liquid on the morphology of systems. We will focus on the dynamics of the capacitive charging, which will provide us with the information of the mechanism of doping/dedopinng on the atomistic scale of the intrinsic capacitance in the presence of ionic liquid.

e-Spect: In silica spectroscopy of complex molecular systems
Theoretical simulations are essential for the microscopic understanding of spectroscopic data that enable the design of biomarkers and materials. Modeling these complex systems requires a combination of molecular dynamics (MD) and quantum mechanics/molecular mechanics (QM/MM) approaches.

Open Space: A tool for space research and communication
OpenSpace is an international open source software development project, with its seat in Norrköping. It is designed to visualize data in astronomy-related research and development.

Feature based exploration of large-scale turbulent flow simulations
This project will enable in-situ detection and tracking of flow structures, which will be used as focus regions to build a multi-resolution description of the data. Interactive visualization methods from volume and flow visualization will be adapted to the new multi- resolution scheme.

Integrated analysis of cardiac function
Recent developments within image-based simulation and imaging of cardiac function can potentially advance diagnosis and treatment optimization of a range of cardiac diseases.

ExABL
A better understanding of the processes in atmospheric boundary layers (ABL) and efficient simulations are required e.g. to improve climate predictions, and simulations of wind parks.

Designing the Next-Generation FFT library
FFT algorithms and software libraries are workforces of scientific computing and data analysis. Most large-scale scientific applications use and rely on FFT libraries, such as FFTW and FFTPACK, which were developed at the end of the Eighties.

Nek5000
We will focus on two aspects in further developing the Nek5000 code within SESSI. First, we will porting Nek5000 to accelerators using OpenACC and CUDA. We will continue the effort of programming Nek5000 for using accelerators to perform batched small matrix matrix multiplication that this the main computational kernel affecting Nek5000 performance. This work will also include optimization with the possibility of using CUDA in combination with OpenACC and improve the efficiency of data movement between host and GPU memories in the GS operator code. This work will be done in collaboration with the EC-funded exascale EPiGRAM-HS project that is led by PDC, and researchers at the Argonne National Laboratory. Second, we will consider new formulations of the compute and communication intensive kernels of Nek5000, including the main communication library gslib. We continue our work on one-sided communication primitives into this kernel via UPC, a PGAS programming system taking advantage of modern network hardware support for efficient one-sided communication. This includes the expertise of Niclas Jansson who developed an initial proof-of-concept of such new software. Features of new languages will also be used to overlap computation and communications by re-organising the flow of the communication.

GROMACS
Molecular simulation has evolved into a standard technique employed in virtually all high-impact publications e.g. on new protein structures. The main bottleneck for scaling in GROMACS is the 3D-FFT used in the particle-mesh Ewald electrostatics (PME). Since PME is very fast, and used by MD codes world wide, it is worth investigating if the communication overhead can be lowered. This is done in collaboration with PDSE (see the 3D-FFT sub-project), as well as a co-design effort with Nvidia for parallelizing the 3D-FFT over GPUs. For extreme scaling, we will also investigate the fast multipole method (FMM) since it has better scaling complexity. A problem was always energy conservation, which is now solved in collaboration with the numerical analysis community, and we will integrate the ExaFMM code of Rio Yokota (Tokyo Tech) into GROMACS.

NumericalAlgebra
We aim to develop new better numerical algorithms for certain problems stemming from data science and machine learning, by using state-of-the-art numerical linear algebra techniques and software and solve the corresponding computationally demanding core problems.

StochasticSimulation
The aim of this project is to develop stochastic simulation methods in Machine Learning along with a corresponding rigorous efficiency analysis.

CausalHealthcare
In this project, we aim to develop machine learning tools for causal discovery and causal inference, along with tools for visualizing large causal structures to a human to increase the interpretability of the structure.

DeepProtein
In this project, we will develop deep learning methods using biological data. In particular, we will address the protein structure prediction problem, which involves predicting the structure from the amino acid sequence, predict interactions with other proteins and peptides, evaluating model qualities and predict amino acid contacts.

DeepClimate
In climate science, we will use datasets collected from existing external projects as well as public datasets to build prediction models that out-perform existing analytical and simulation models.

DeepTurbulence
In this project, we propose to develop and use generic Deep Learning techniques that are able to model physical (simulated and/or measured) dynamics.

Variational approximations in the medical sciences
In this new sub-project we will develop machine learning models and tools for Gaussian variational approximations (GVAs) and apply those models to health applications.

Cancer screening – natural history, prediction and microsimulation
We will continue work on natural history modelling for cervical, breast and prostate cancer. Methods include HPC-intensive calibrations of simulation likelihoods using Bayesian methods and optimisation procedures for expensive or imprecise objective functions (Laure, Jauhiainen at AstraZenenca, Uncertainty Quantification with SeRC-Brain-IT). We will investigate a computational framework for storage and analysis of m icro-simulation experiments for calibration and prediction (Laure, Dowling).

Medical image analysis and deep learning, with applications to prostate biopsies and mammograms
The ability to digitise large quantities of medical images together with recent progress in the area of deep learning and stochastic modelling of highly structured systems offers an opportunity to change and improve diagnostic procedures for screening.

Trial design for prediction
Experimental design is an under-appreciated aspect of data science, where better design leads to more efficient parameter estimation and possibilities to address causality. We will contribute to two new studies:
- the STHLM3-MRI study to assess the combination of the S3M test with magnetic resonance imaging (MRI), and
- the ProBio randomised treatment trial for men with metastatic prostate cancer.

Translational bioinformatics: Statistical learning for patient stratification
Translational bioinformatics was introduced into eCPC during 2014, when formal collaborations with eSSENCE and the SciLifeLab Clinical Diagnostics facility in Uppsala were also initiated.

Brain-like approach to Machine Learning
The main aim of this project is to advance the development of hierarchical brain-like network architectures for holistic pattern discovery drawing from the computational insights into neural information processing in the brain in the context of sensory perception, multi-modal sensory fusion, sequence learning and memory association among others

Uncertainty quantification and sensitivity analysis in brain modelling
Uncertainty quantification and sensitivity analysis are important aspects of computational modelling, due to the need to assess the validity and precision of model predictions.

Brain network architecture and dynamics of short- and long-term memory
In this project we intend to study cortical network phenomena accompanying brain plasticity effects relevant to short- and long-term memory processes. The overarching aim is to enhance e-science approaches for studying brain networks developed at KTH and KI, and inject corresponding informatics workflows into the environments at SUBIC. PH plans to advance an existing spiking and non-spiking large-scale neural network models to simulate memory phenomena in close collaboration with AL.




