Selection Concepts for Complex Molecular Structures
Research focus area – Multi-scale visualization
Application – Molecular dynamics simulations for conformation analysis
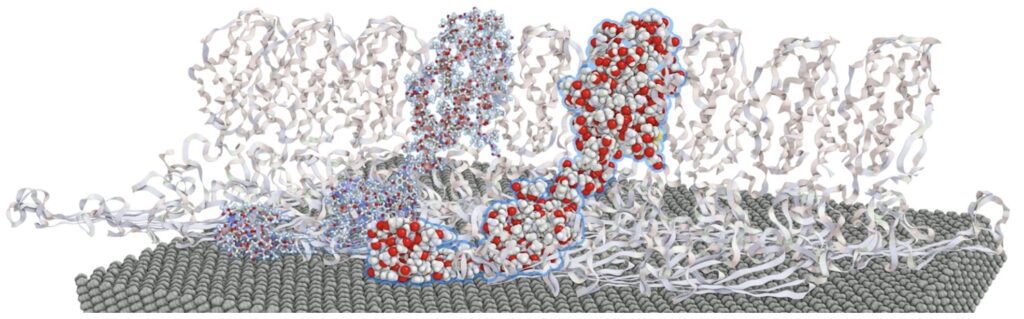
“It is through the interactive manipulation of a visual interface – the analytic discourse – that knowledge is constructed, tested, refined, and shared.” [Pike 2009]. This is also true for molecular dynamics (MD) simulation data, e.g., for studying bindings between ligands and molecules, an essential part of nanotechnologies and life sciences. Therefore, a selection framework that can cope with dense dynamically changing visual representations has been developed. It is integrated into a visual analytics tool mostly targeted toward expert users from the MD domain. It combines a large set of selection paradigms in a flexible way while maintaining a simple and intuitive interface with immediate visual feedback. This includes text-based queries, temporal and property-based filtering, brushing functionality, and morphological and structure-based growing and shrinking operations combined using Boolean operators. Typical use cases are the selection of structures to track properties overtime or the specification of visual representation styles to highlight events for communication purposes.
Image: Growing an initial selection of two residues along with covalent bonds in a protein fibril.
Reference:
Skånberg et al. MolFind – Integrated Multi-Selection Schemes for Complex Molecular Structures, Eurovis workshop on molecular visualization, 2019.




