Binding sites for luminescent amyloid biomarkers from non-biased molecular dynamics simulations
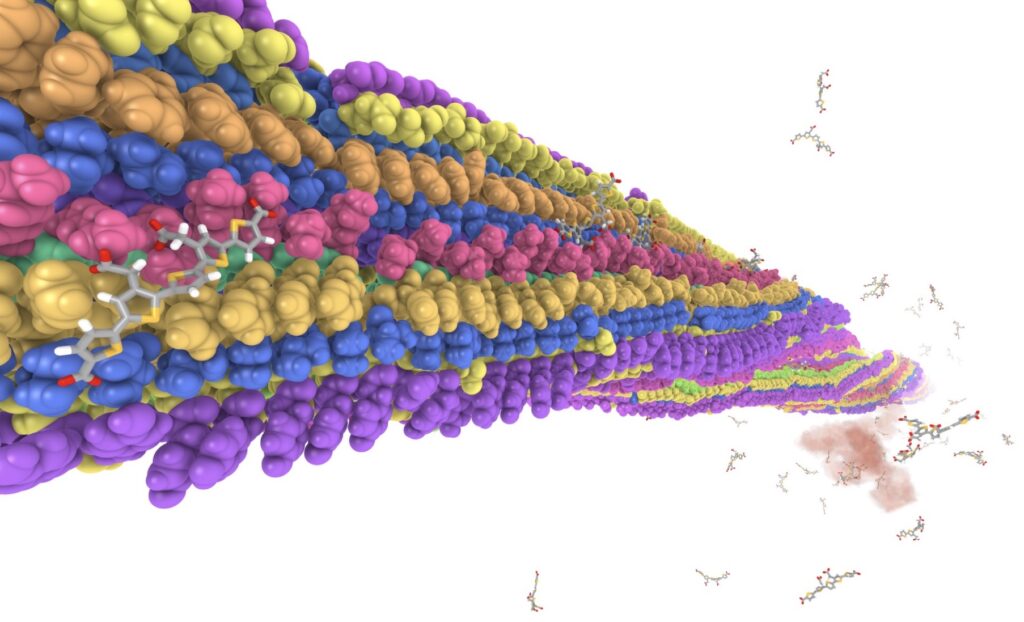
A very stable binding site for the interaction between a pentameric oligothiophene and an amyloid- beta(1–42) fibril has been identified by means of non-biased molecular dynamics simulations. In this site, the probe is locked in an all-trans conformation with a Coulombic binding energy of 1200 kJ/mol due to the interactions between the anionic carboxyl groups of the probe and the cationic amino groups in the lysine side chain. Upon binding, the conformationally restricted probes show a pronounced increase in molecular planarity. This is in line with the observed changes in luminescence properties that serve as the foundation for their use as biomarkers.
König, C., Skånberg, R., Hotz, I., Ynnerman, A., Norman, P., & Linares, M. (2018). Binding sites for luminescent amyloid biomarkers from non-biased molecular dynamics simulations. Chemical Communications, 54(24), 3030–3033.
https://doi.org/10.1039/C8CC00105G
This project represents a cornerstone in the European Innovative Training Network (ITN) that is formed between the following nodes in Stockholm, Heidelberg, Lyngby, Trondheim, Odense, Paris, Munich, and Pisa. The ITN runs for the period of 2018–2021 with a budget of 3.75 MEuro and is titled Computational Spectroscopy in Natural Sciences and Engineering (COSINE). Further details may be found at https://bit.ly/itncosine.




