Ensemble parallelism in GROMACS
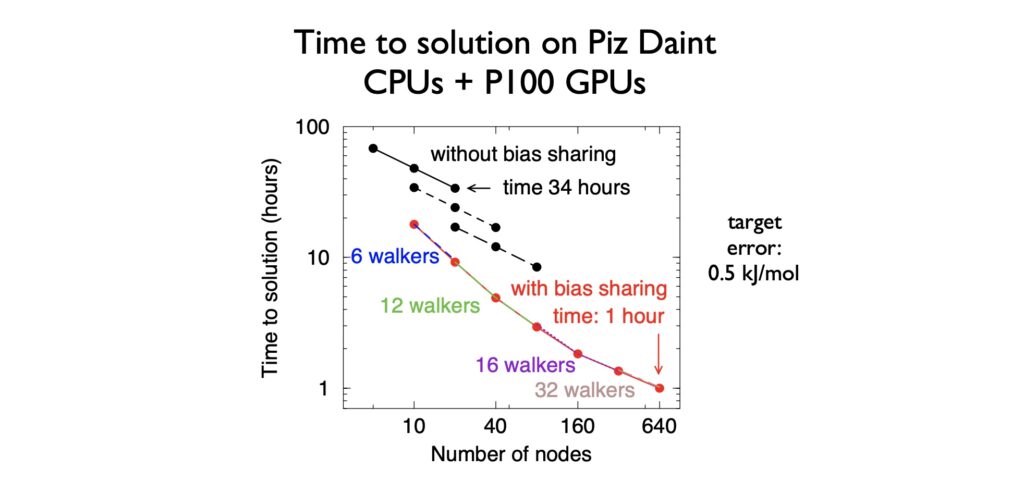
Scaling individual atomistic simulations of bio-molecules is a strong scaling problem, as the molecules have fixed size. To reach the exascale, this needs to be combined with ensemble parallelism. Since ensemble properties are always what one wants to calculate, this is a natural fit. As a showcase, we have applied the ensemble parallel accelerated weight histogram method to a small DNA system. Many walkers are used to accurately map out the free-energy landscape of opening of a DNA base pair, which is an essential step in any function of DNA. The time to solution was reduced by a factor of 34 using 640 GPU-accelerated notes of Piz Daint at CSCS in Switzerland.




