Characterization of molecular motion in Cryo-EM image reconstruction
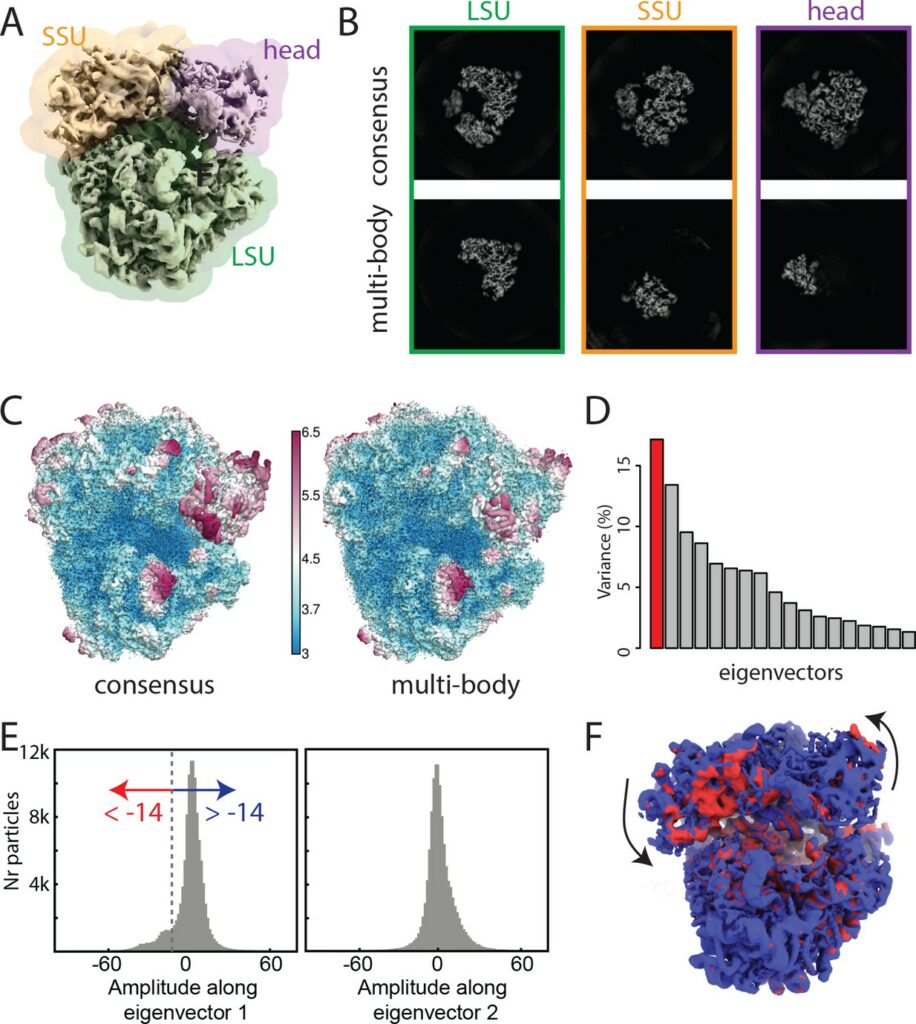
In collaboration with the Laboratory of Molecular Biology (Cambridge, UK), the SeRC Molecular Simulation community has continued their work on new reconstruction algorithms for cryo-electron microscopy. During 2017, we have particularly introduced new methods that allow different parts of large molecules to move relative to each other during (rigid) multi-body optimization. This solves the common problem where e.g. the different proteins in a ribosome move slightly relative to each other (which has previously led to lost resolution), and both improves the local resolution as well as provides information about how the molecules are moving in the sample by extracting principal components of the data. This work has been introduced in the widely used RELION code we develop together, it resulted in a high-impact eLife publication [Nakane et al, eLife 2018;7:e36861]. In addition to our ongoing close RELION & GROMACS collaborations with NVIDIA inc., we have also engaged in a new collaboration with INTEL inc. to optimize cryo-EM reconstruction for traditional processors, and this work was awarded the prize for best parallel programming at the Intel HPC Developer Conference 2017.




