Brain-IT Highlight: Workflows for the estimation of model parameters
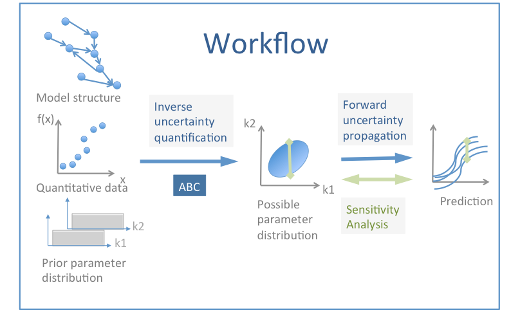
When modeling subcellular signaling pathways, experimental data are integrated into a precise and structured framework from which it is possible to make predictions that could be tested experimentally. However, the quantitative experimental data that are used for the building of pathway models are often sparse as compared to the size and complexity of the modelled system, and the translation of these data into dynamical models leaves numerous uncertainties in parameter values. We have together with the eCPC community developed a workflow for building and testing intracellular signaling models and for the quantification of model parameter uncertainty, and its propagation to predictions. This workflow is applied on a model describing calcium (Ca)-dependent activation of Calmodulin (CaM), Protein phosphatase 2B (PP2B) and Ca/CaM-dependent protein kinase II (CaMKII).
References
Eriksson O, Jauhiainen A, Sasane SM, Nair AG, Sartorius C and Hellgren Kotaleski J (2016) Statistical uncertainty and sensitivity analysis of intracellular signaling models – through approximate Bayesian computation and variance based global sensitivity analysis, INCF Congress abstract, htttp://www.frontiersin.org/10.3389/conf.fninf.2016.20.00014/event_abstract




