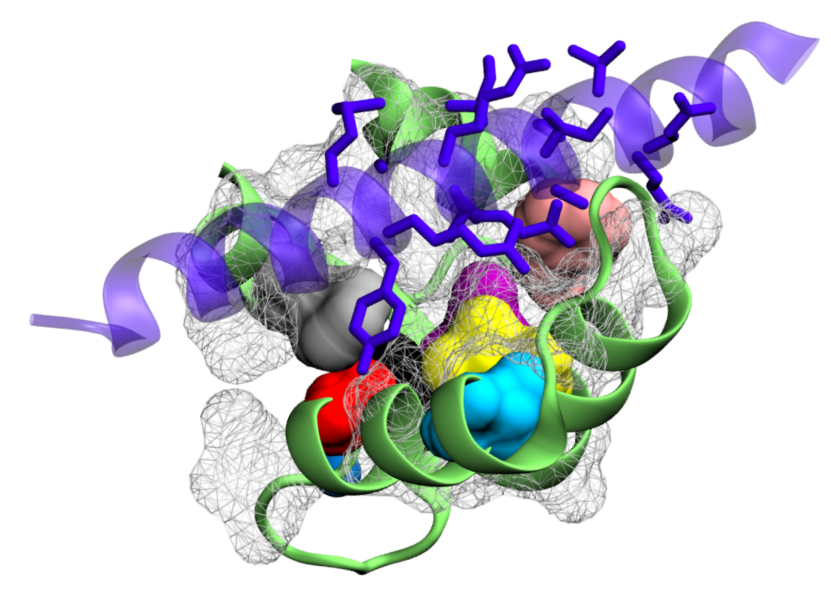
Calmodulin is a ubiquitous calcium sensor that confers calcium sensitivity to many cellular partners. How calmodulin can bind a large number of targets while retaining some selectivity, a phenomenon called promiscuous selectivity, is a fascinating question that we address with molecular dynamics simulations conducted with GROMACS. After sampling the conformational landscape of calmodulin in the presence and absence of calcium ions, we use smart data analysis to discover stable states and to verify their compatibility with protein binding. Binding calcium indeed modifies the accessible hydrophobic surface of the two calmodulin lobes and allows for deeper binding. We further identify that the long-ranged electrostatic interactions of the charged residues of one lobe are prone to initiating binding, while the short-ranged interactions of hydrophobic residues in the other lobe provides binding selectivity. The two lobes thus join forces to allow promiscuous selectivity.
REFERENCES:
Inference of Calmodulin’s Ca2+-Dependent Free Energy Landscapes via Gaussian Mixture Model Validation AM Westerlund, TJ Harpole, C Blau and L Delemotte, J. Chem. Theor. Comp. 2018 DOI: 10.1021/acs.jctc.7b00346
Effect of Ca2+ on the promiscuous target-protein binding mechanism of calmodulin A Westerlund, L Delemotte, PloS Comp. Biol. 2017 DOI: 10.1371/journal.pcbi.1006072