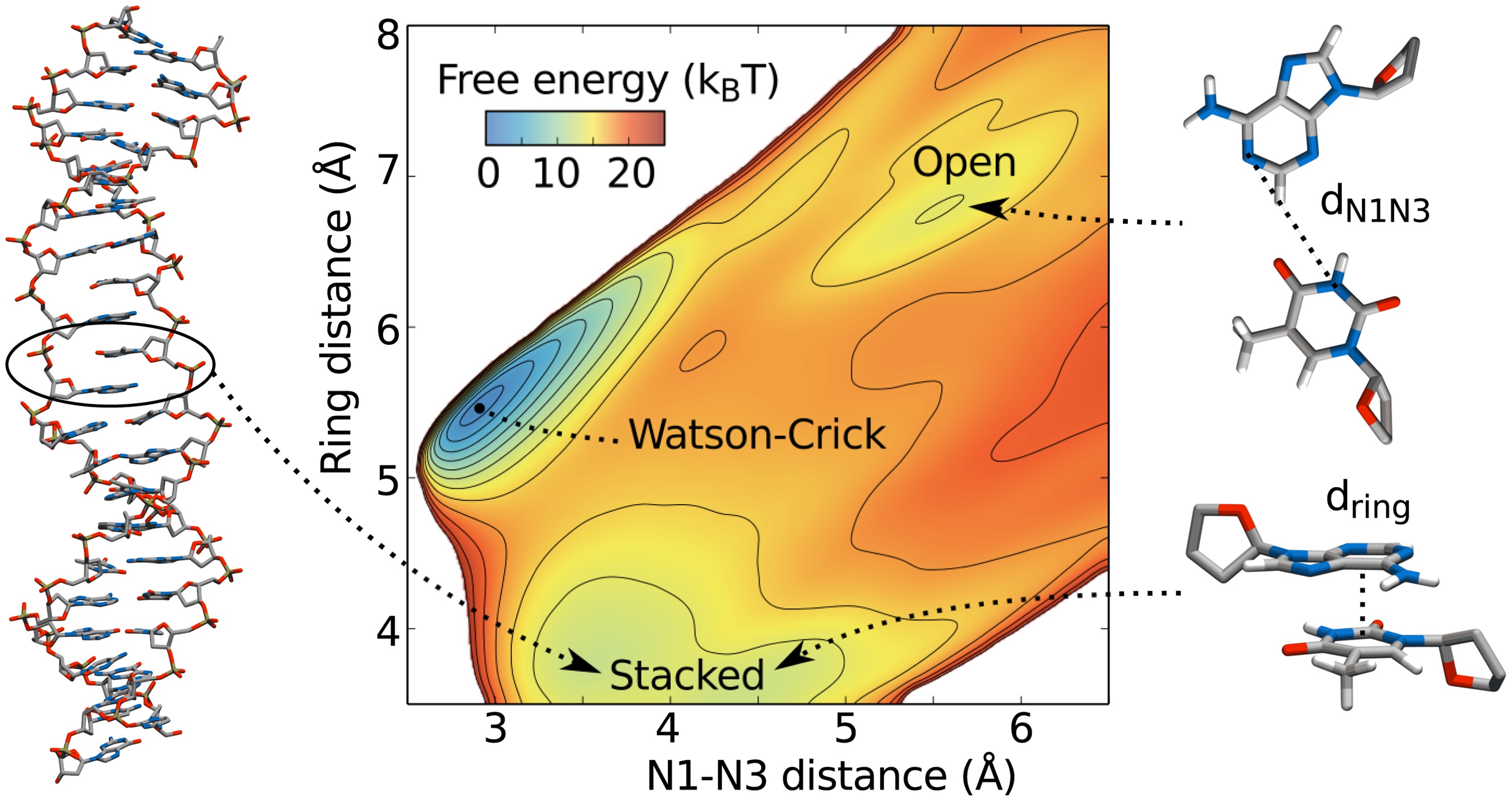
Molecular dynamics is a powerful technique to study the behavior of molecules, and in particular bio-molecules such as proteins and DNA. However, the time scales of biologically important events are often much longer than what can be reached with simulations. Usually this is caused by free-energy barriers along the pathway, which are only crossed after many failed attempts. We have developed a technique called the Accelerated Weight Histogram (AWH) method, which exponentially accelerates crossing of barriers. Unlike many other methods, AWH has no critical parameters and will accelerate sampling along suitably chosen reaction coordinates in an automated fashion. In this project we investigated DNA base-pair opening, which occurs in all biologically relevant processes, since bases need to be exposed to be recognized or modified. Base pair opening occurs on the millisecond time scale, so accelerated sampling is required to be able to sample opening in less than a microsecond. AWH is available to anyone through inclusion in the 2018 release of the popular GROMACS open source simulation package.
Sequence dependency of canonical base pair opening in the DNA double helix
V. Lindahl, J. Lidmar and B. Hess
PLOS Computational Biology, 13 (4), e1005463 (2017)
Accelerated weight histogram method for exploring free energy landscapes
V. Lindahl, J. Lidmar and B. Hess
J. Chem. Phys. 141, 044110 (2014)